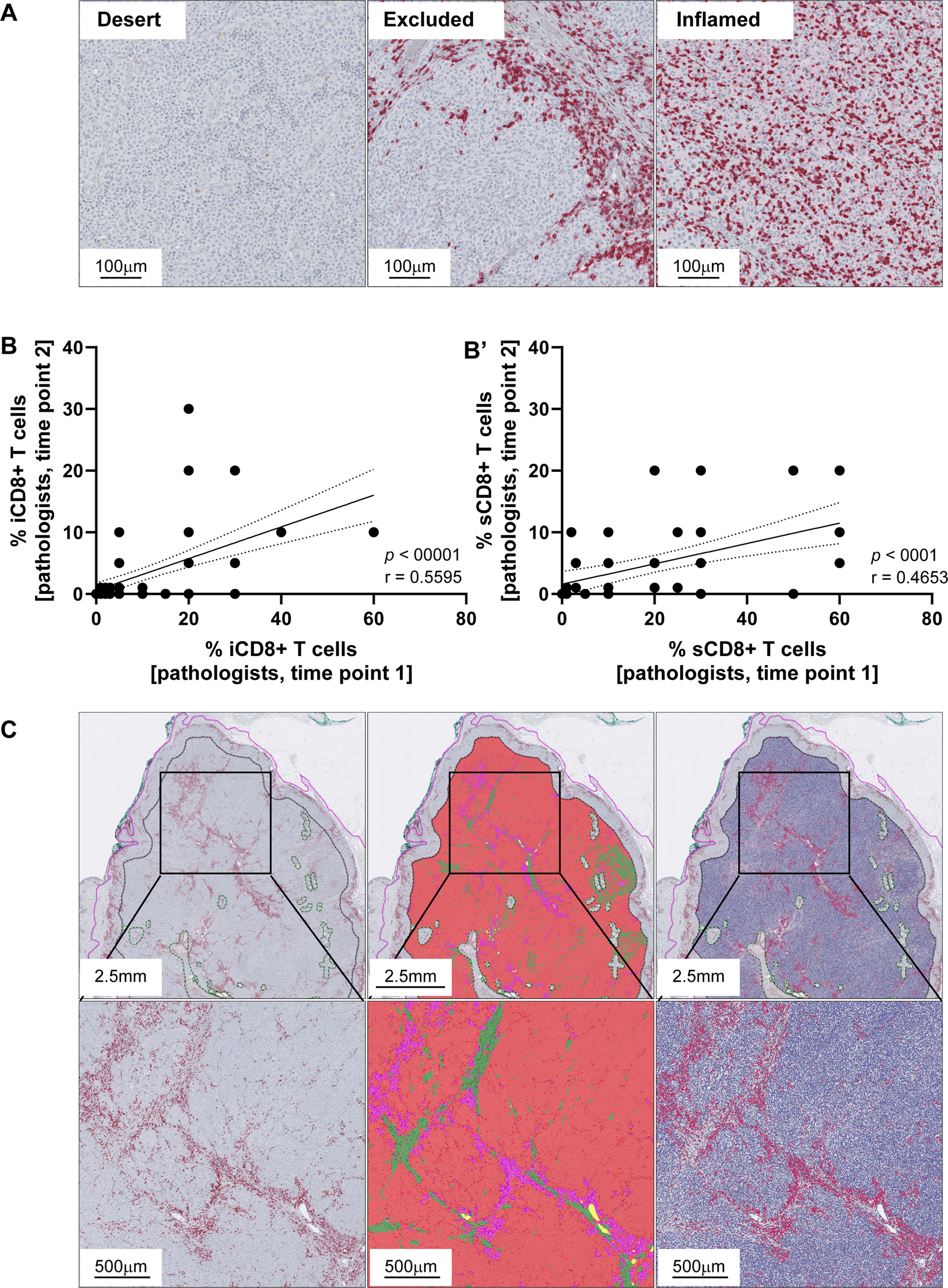
Establishing standardized immune phenotyping of metastatic melanoma by digital pathology | Laboratory Investigation

Figure A1: Comparison of inferred mutation clusters when 25, 50 or 100... | Download Scientific Diagram
Identification of Constrained Cancer Driver Genes Based on Mutation Timing | PLOS Computational Biology
Identification of Constrained Cancer Driver Genes Based on Mutation Timing | PLOS Computational Biology

PDF) Computational Strategies to Combat COVID-19: Useful Tools to Accelerate SARS-CoV-2 and Coronavirus Research | Anne-Christin Hauschild - Academia.edu
We gratefully acknowledge the following Authors from the Originating laboratories responsible for obtaining the specimens, as w
Identification of Constrained Cancer Driver Genes Based on Mutation Timing | PLOS Computational Biology
Identification of Constrained Cancer Driver Genes Based on Mutation Timing | PLOS Computational Biology
Inferring ongoing cancer evolution from single tumour biopsies using synthetic supervised learning | PLOS Computational Biology

Metagenomic Investigation of Viral Communities in Ballast Water | Environmental Science & Technology

Progress Identifying and Analyzing the Human Proteome: 2021 Metrics from the HUPO Human Proteome Project | Journal of Proteome Research